National Center for Biotechnology Information , U. Ryuta Muromoto 21 Estimated H-index: Stem Kernels The stem kernel is a kernel function for structural RNAs to measure a kind of similarity between a pair of RNA sequences from the viewpoint of secondary structures. The equilibrium partition function and base pair binding probabilities for RNA secondary structure. RNA secondary structure prediction without physics-based models. Alternative approach is based on probabilistic frameworks, including stochastic context-free grammars SCFGs , which can model RNA secondary structures without pseudoknots 3. 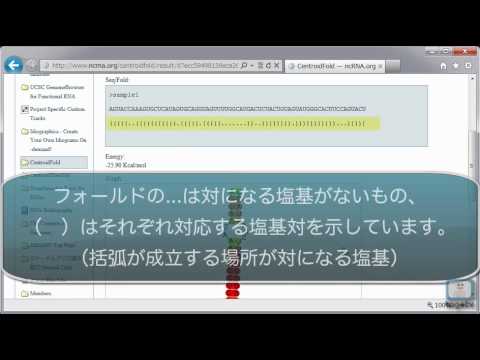
| Uploader: | Arashiramar |
| Date Added: | 24 March 2013 |
| File Size: | 58.82 Mb |
| Operating Systems: | Windows NT/2000/XP/2003/2003/7/8/10 MacOS 10/X |
| Downloads: | 19212 |
| Price: | Free* [*Free Regsitration Required] |
PDF version of the graph representation is also available.
CentroidFold: a web server for RNA secondary structure prediction
Stem Kernels The stem kernel is a kernel function for structural RNAs to measure a kind of similarity centroidtold a pair of RNA sequences from the viewpoint of secondary structures. Evaluation of several lightweight stochastic context-free grammars for RNA secondary structure prediction.
CentroidFold CentroidFold based on a generalized centroid estimator is one of the most accurate tools for predicting RNA secondary structures.
The C entroid F old web server can be accessed on http: It is based on a dynamic programming algorithm from applied mathematics, and is much more efficient, faster, and can fold larger molecules than procedures which have appeared up to now in the biological literature. Recent studies have shown that the methods for predicting secondary structures of RNAs on the basis of posterior decoding of the base-pairing probabilities has an advantage with respect to prediction accuracy over the conventionally utilized minimum free energy methods.
As it is well known, statistical decision theory shows that maximum likelihood and related estimators use data only to identify the single most probable solution. Vienna RNA secondary structure server. Current Centroiefold drugs are split into two groups: RNA secondary structure prediction without physics-based models Chuong B.
Softwares – Bioinformatics Tools and Databases for Functional RNA Analysis
The major advantage of this server is that it employs our original C entroid F old software as its prediction engine which scores the best accuracy in our benchmark results. Fdur Fdur computes sufficient statistics of phylogenetic tree models Rentropy computes entropy, mean energy, and variation centroidcold energy from the Boltzmann distribution of RNA secondary structures Rchange Rchange computes energy changes of RNA secondary structures after base mutations.
Here, we identified numerous nove Centroid estimation in discrete high-dimensional spaces with applications in biology. CentroidFold based on a centroidcold centroid estimator is one of the most accurate tools for predicting RNA secondary structures.
CentroidFold: a web server for RNA secondary structure prediction
Joseph Fogerty 2 Estimated H-index: Abstract The accurate prediction of RNA secondary structure from primary sequence has had enormous impact on research from the past forty years. RNA secondary structure prediction using stochastic context-free grammars.
In this article, we introduce a web application of C entroid F old with a very simple interface. The server quickly responses with a prediction result. These should be included in structural analyses to give improved results. Kengo Sato 16 Estimated H-index: The difference between them is only in the gain functions: Do 31 Estimated H-index: Journal List Nucleic Acids Res v. Unlike physics-based methods, which rely on thousands of experimentally-measured thermodynamic parameters, SCFGs use fully-automated statistical learning algorithms to derive model parameters.
The server quickly responses with a prediction result. Accordingly, unless this one solution so dominates the Support Center Support Center. Hofacker 58 Estimated H-index: This article has been cited by other centroidfols in PMC. KozlowskiKristian M.
However, the number of free parameters in an integrated model for consensus RNA structure prediction can become untenable if See 10 for more ecntroidfold. HirutaShin-ichi Hiruta.


Комментарии
Отправить комментарий